Welcome to the Annotation of the Transcriptional Cancer Atlas!¶
Version: 0.1.0
Last change: May 07, 2022
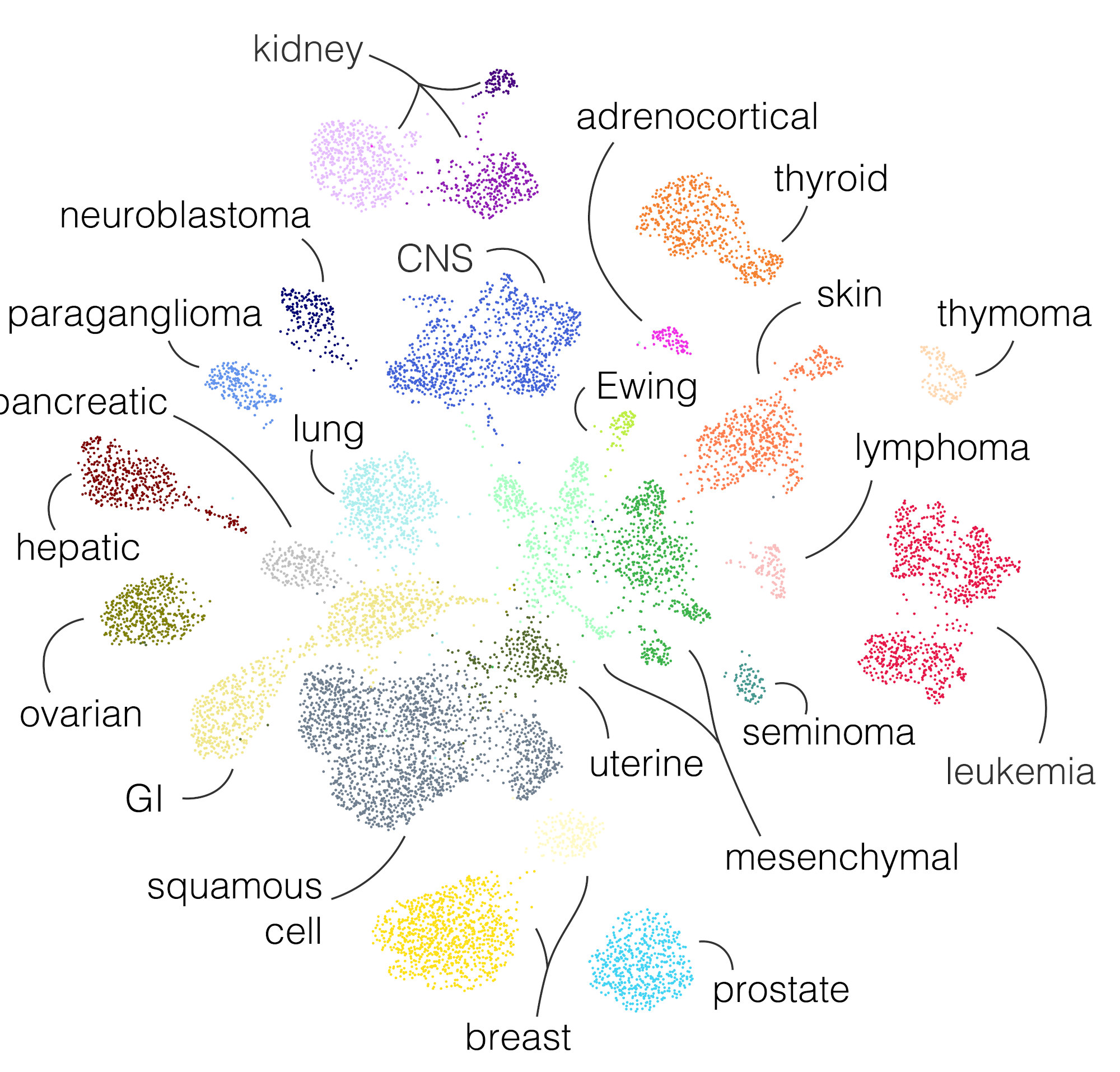
This website contains a detailed description of the classes available in the transcriptional cancer atlas, a curated resource of gene expression data clustering from the Hospital for Sick Children. The atlas was produced with RACCOON, a scale-adaptive clustering algorithm, and serves as a target to the OTTER classifier.
The data was gathered from different sources, including the TreeHouse Childhood Cancer Initiative, TCGA, TARGET , GTEx, and St. Jude Children’s Research Hospital.
The classes have been manually annotated. Each one has been characterized based on its most relevant transcriptional properties and, where available, clinical and genomics information.
This website is still a work in progress. We are striving to cover all subtypes currently available in the atlas, please check back in the future if what you are looking for is still not available. For questions, comments or suggestions, please Contact us. See also our Frequently Asked Questions section.
IMPORTANT NOTE: The annotation is updated periodically, make sure you are looking at a version that is matching the classifier used in your project.
If you are using this annotation data or related software, please cite:
F. Comitani, J. O. Nash, S. Cohen-Gogo, A. Chen Schwertschkow, T. T. Wen, A. Maheshwari, B. Goyal, E. S. L. Tio, K. Tabataei, L. Brunga, J. E. G. Lawrence, P. Balogh, A. Flanagan, S. Teichmann, V. Ramaswamy, J. Hitzler, J. Wasserman, R. A. Gladdy, B. C. Dickson, U. Tabori, M. J. Cowley, S. Behjati, D. Malkin, A. Villani, M. S. Irwin and A. Shlien, “Multi-scale transcriptional clustering and heterogeneity analysis reveal diagnostic classes of childhood cancer” (under review).
Copyright ©2022 Federico Comitani, Josh O. Nash and Adam Shlien.
RESEARCH USE ONLY: This website is intended for research purposes only. These analyses were not performed for the purposes of diagnosis, prophylaxis, or treatment.