N000 Healthy Normal Tissue¶
Version: 0.1.0
Last change: May 07, 2022
We included in our dataset 1735 RNASeq samples from 31 locations and 52 tissue types within the the GTEx project, [GTEx2013], [Carithers2015]. Samples from cell cultures (such as fibroblasts and lymphblastoids) were not included in this work.
The resulting hierarchy of clusters is shown in Fig. NORM1. In a number of cases we observe the merging of tissue types common functional features [Uhlén2015]. To help with the annotation, we looked at histological report shared, which revealed considerable variability of cell types across samples from the same organ.
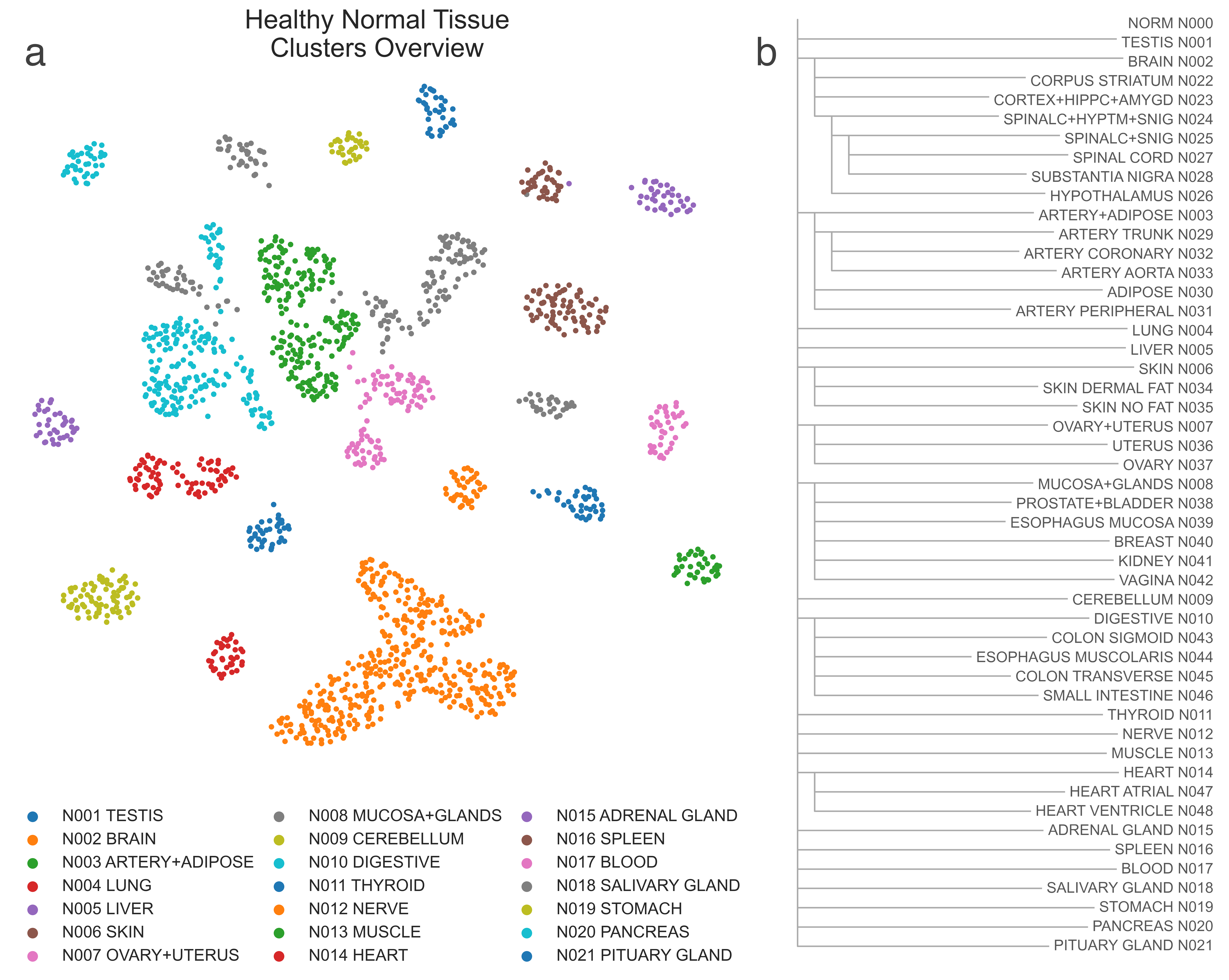
LEU1: A, 2-dimensional UMAP projection of healthy normal tissue samples by gene expression. Subtypes at the first level of the hierarchy are shown with different colours. B, the list of all healthy normal tissue subtypes identified and their hierarchical relationship.¶
Samples from the digestive tract are found in N010 DIGESTIVE (n = 192) and branch out into N043 COLON SIGMOID (n = 54),
N044 ESOPHAGUS MUSCOLARIS (n = 82), containing both muscolaris and esophageal junction samples),
N045 COLON TRANSVERSE (n = 27) and N046 SMALL INTESTINE (n = 29), although a degree of overlap is observed
across all these groups. N044 attracts samples from other organs
with a majority of smooth muscle tissue.
The database contains only 10 bladder samples (the only ones available on
GTEx at the time of this work), five of which are found within this group.
The remaining half cluster with prostate tissue N038 PROSTATE+BLADDER (Prostate or bladder)` (n = 37),
but could be possibly be further separated if the population were to be increased.
This last group is part of a more general supercluster of mostly gland and hormonal
regulatory tissues (N008 MUCOSA+GLANDS, n = 161), esophagus mucosa and submucosa glands
(N039 ESOPHAGOUS MUCOSA, n = 39, including two salivary gland samples with a majority of mucosa and stroma),
breast (ducts and glands), fallopian and endocervix tissues (N040 MAMMARY, n = 26), kidney cortex, n = 27) vaginal epithelium and mucosa, and ectocervix (N042 VAGINA, n = 32). As observed before,
the borders between female reproductive organs tissues also tend to overlap, depending on the ratio between mucosa,
muscle, or other tissue types within the sample.
Cervix and vaginal tissues are also found to a lesser degree in N007 OVARY+UTERUS (n = 92), which is then split into
N036 UTERUS (n = 52), and N037 OVARY (n = 40) at the next iteration.
Occasionally, the separation is orthogonal to provided labels.
N005 SKIN (n = 75) samples are not grouped by sun exposure, but rather by the presence or absence of any dermal fat:
N034 SKIN DERMAL FAT (n = 51) which also includes a single sample labelled as subcutaneous adipose tissue and
N035 SKIN NO FAT (n = 24) respectively.
Heart tissue samples are found in N013 HEART (n = 74), which then splits by location into
N047 HEART ATRIAL (n = 36) and
N048 HEART VENTRICLE (n = 38).
Arteries are initially clustered with adipose tissues in N002 ARTERY+ADIPOSE (n = 203), possibly due to their fat component,
and are then separated in peripheral N031 ARTERY PERIPHERAL (n = 34), and coronary and aorta
N029 ARTERY TRUNK (n = 78) which are then split in their specific types, N032 ARTERY CORONARY (n = 42) and
N033 ARTERY AORTA (Artery aorta)` (n = 36) respectively. N030 ADIPOSE (n = 91) is a relatively big group, containing fat tissues from different body parts.
It includes a majority of visceral and subcutaneous adipose tissue, but also breast and a few gastroesophageal junction samples.
N032 also contains a few samples from tibial arteries, and, more interestingly two salivary gland and a vagina sample.
According to the histological analysis, the salivary gland samples are missing glands and mostly made of fibromuscular tissue,
while the vagina sample contains almost exclusively fibrovascular stroma, further supporting the importance of proper expression
profiling for normal samples too. This is particularly important in the context of tumor-normal matching to build a background population.
Finally, most central nervous system regions overlap considerably, except for Cerebellum samples,
which are cleanly grouped together in N009 CEREBELLUM (n = 74), which also includes a single mislabelled cortex sample
(as confirmed by the histological report).
The remaining samples are found in N001 BRAIN (n = 398) and its subclusters:
N022 CORPUS STRIATUM (n = 96) contains tissues from the nucleus accumbens,
the caudate nucleus and putamen; hippocampus,
amygdala, and cortex, tend to overlap significantly and are all grouped in the single biggest cluster
N023 CORTEX+HIPPC+AMYGD (n = 193), which couldn’t be further separated;
and finally N024 SPINALC+HYPTM+SNIG (n = 109)
comprises instead spinal cord, hypothalamus and substantia nigra samples. The latter further splits however into
N026 HYPOTALAMUS (n = 30), and then again N027 SPINAL CORD (n = 51) and
N028 SUBSTANTIA NIGRA (n = 27), although a
good degree of overlap is observed between the last two. Although N028 contains the majority of substantia nigra
samples, this tissue type is observed across most of the other clusters too.
Bibliography¶
- Carithers2015
Carithers, L.J., Ardlie, K., Barcus, M., et al. 2015. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreservation and biobanking 13(5), pp. 311–319.
- GTEx2013
GTEx Consortium 2013. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 45(6), pp. 580–585.
- Uhlén2015
Uhlén, M., Fagerberg, L., Hallström, B.M., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347(6220), p. 1260419.